Single-cell Sequencing refers to analyzing the sequence information of each single cell through NGS (Next Generation Sequencing), so as to reveal the cellular differences between cells and their functions in the microenvironment with higher resolution.
Why do single cell sequencing
More than a decade of single-cell sequencing has made our understanding of the world more precise. Our traditional sequencing method is carried out at the tissue level or multi-cell level, and the final data is actually the average of multiple or multiple types of cells, but the information of cell heterogeneity is not clear. Single-cell sequencing, on the other hand, can obtain genetic information at a finer level of individual cells, thus revealing the gene structure and gene expression status of each cell, and reflecting the genetic information of heterogeneous and rare cells that cannot be obtained by conventional sequencing. Therefore, single-cell sequencing has been called the "microscope" of NGS.
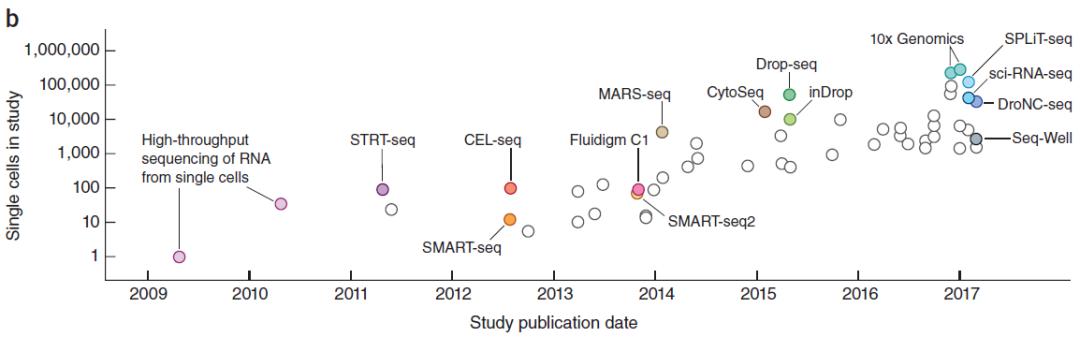
Figure 1. Development history of single cell sequencing
Difficulties in single cell sequencing technology
The biggest limitation of single-cell sequencing lies in the content of nucleic acid to be measured. For example, the content of DNA or RNA in a cell is only at the Picograms level, far less than the minimum input requirements of existing library building kits. Therefore, the nucleic acid molecules in a single cell must be amplified first, with as few technical errors as possible, in order to carry out subsequent sequencing and other studies. Second, high-throughput single-cell sequencing requires increased capture efficiency and sensitivity. In addition, the acquisition of rare samples and cell separation of hard-to-dissociate tissues are also great challenges in sample processing.
Several major single-cell sequencing techniques
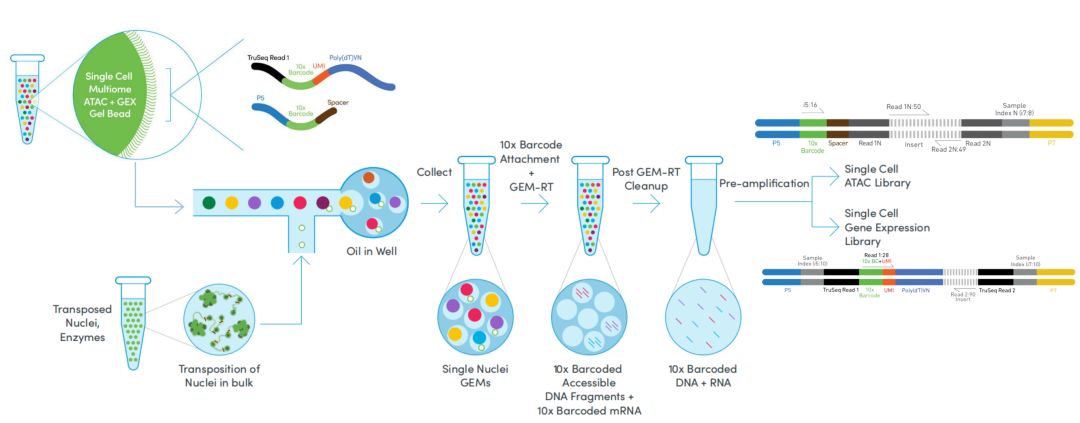
◎ 10× Genomics based on microfluidic droplets
The Chromium system at 10× Genomics uses microfluidic technology to automatically assign cells to 100,000 to 1,000,000 microresponse systems, each containing a specific barcode sequence. The barcode information gel beads (GEMs) are first mixed with a mixture of samples and enzymes and then combined with an oil surfactant solution located in a microfluidic "double cross" junction; The GEMs flow into the reservoir and are collected. The gel bead dissolves and releases the barcode sequence, and the sample is labeled. Finally, the products containing barcode information in each droplet were mixed and the standard sequencing library was constructed.
The advantages of 10× Genomics are high cell fluxes, short library building cycles, ease of operation, and high capture efficiency. The disadvantage is that only 3 'end transcript information can be obtained, and the sample requirements are high.
Figure 2. Principle of 10× Genomics sequencing
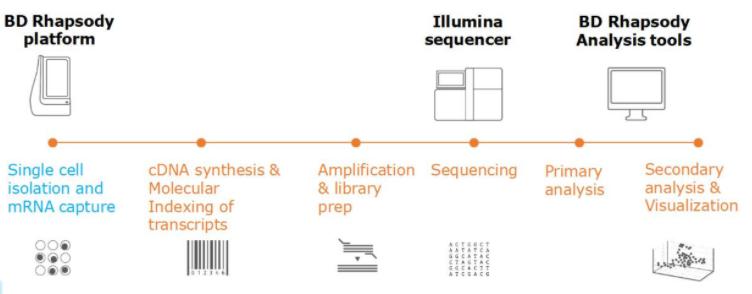
◎ BD Rhapsody based on microplates
The BD Rhapsody technology enables single-cell capture and molecular labeling of mRNA transcript on magnetic oligonucleotide bar-coded microspheres using card chips, which are then incorporated into a single tube for cDNA amplification and library construction. It can be used for automatic sorting, amplification and library construction of 100~10,000 cells. In addition, the technique uses high-quality antibodies with oligonucleotides (AB-Oligos), which have an antibody-specific bar code that allows cells to obtain both transcriptome and protein expression at the single-cell level after ab-OligOS labeling.
Compared to 10× Genomics, BD Rhapsody has some advantages in capture efficiency and effectiveness.
Figure 3. Principle of BD Rhapsody sequencing
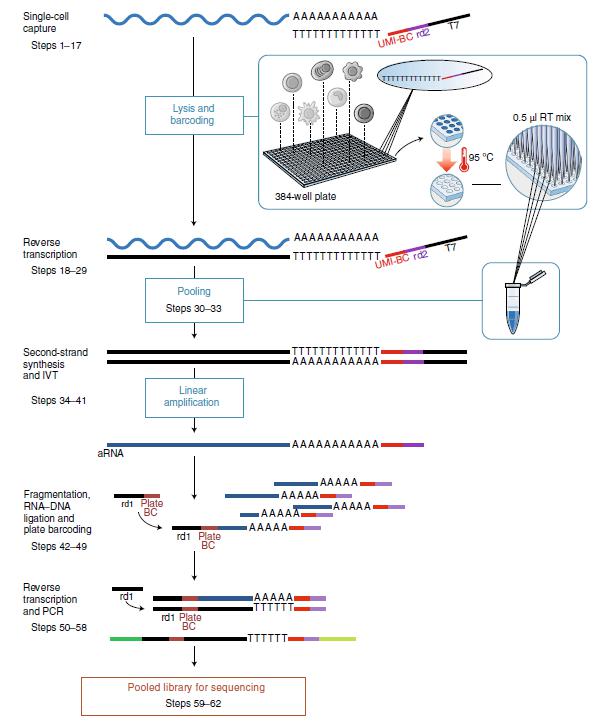
◎ Smart-SEQ technology based on PCR amplification
Smart-seq (Switching mechanism at 5 'end of the RNA transcript) is a landmark technique. After the cell is lysed, RNA is hybridized with primers containing Oligo (dT). A few template-free C nucleotides are then added to generate the first strand. This Poly (C) pendant is only added to the full-length transcript. The oligonucleotide primer was hybridized with poly(C) highlight to synthesize the second chain. The full-length cDNA was amplified by PCR to obtain nack-grade DNA. PCR products can be purified for sequencing. Later, some scholars improved the technology, and the new scheme used clna (LNA), higher concentration of MgCl2 and betaine. It eliminates the need for purification steps and can greatly increase yield.
Smart-seq detected more transcripts than 10× Genomics, but lacked the ability to achieve chain specificity due to sequencing reads, and was not suitable for analysis of non-Poly (A) Rnas due to its selectivity to polyadenylated Rnas.
Figure 4. Principle of Smart-SEQ sequencing
Mars-seq technology based on linear amplification
By connecting T7 promoter to oligo dT primer, IVT (in vitro transcription) can be activated after cDNA synthesis. Mars-seq 2.0 is based on mars-SeQ. A process that integrates index sorting and a large number of parallel single-cell RNA sequencing methods.
Figure 5. Principle of Mars-SEq sequencing
Development prospect of single cell sequencing technology
In 2013, single-cell sequencing technology was named technology of the Year by Nature Methods. That same year, Science ranked single-cell sequencing at the top of its list of six areas to watch this year. At the same time, with the development and update of technology, single-cell sequencing technology has developed from the high cost, low throughput and low automation in the early stage to the current low cost, high throughput and high automation, and its clinical application has developed rapidly. It can quickly determine the precise gene expression pattern of thousands of cells and analyze the genetic heterogeneity of each cell. Single cell sequencing technology has only been developed for more than 10 years, playing an important role in neurobiology, tumor research, prenatal gene diagnosis and other fields, and has become the focus of scientific research.
reference
< drop down view >
【1】. Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods[J]. Molecular cell, 2017, 65(4): 631-643. e4.
【2】. Method of the Year 2013. [J]. Nat Methods ,2014,11:1.
【3】. The biology of genomes. Single-cell sequencing tackles basic and biomedical questions.[J] .Science, 2012, 336: 976-7.
【4】. Navin Nicholas E, Delineating cancer evolution with single-cell sequencing.[J] .Sci Transl Med, 2015, 7: 296fs29.
【5】. Svensson Valentine,Vento-Tormo Roser,Teichmann Sarah A, Exponential scaling of single-cell RNA-seq in the past decade.[J] .Nat Protoc, 2018, 13: 599-604.
【6】. Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014 Jan;9(1):171-81. doi: 10.1038/nprot.2014.006. Epub 2014 Jan 2. PMID: 24385147.
【7】. Keren-Shaul H, Kenigsberg E, Jaitin DA, David E, Paul F, Tanay A, Amit I. MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat Protoc. 2019 Jun;14(6):1841-1862. doi: 10.1038/s41596-019-0164-4. Epub 2019 May 17. PMID: 31101904.